Research
Major current research strengths include
- Immunity to influenza and other respiratory pathogens, supported by the NYICE contract (Topham, Sant, Sangster, Mosmann, Quataert)
- Mechanisms of neutrophil and T cell migration (Kim)
- In vivo imaging of immune function supported by the Imaging PPG (Kim, Topham)
- Optogenetics and cancer therapy (Kim)
- T cell signaling and differentiation (Miller, Mosmann)
- Antigen presentation (Sant, Livingstone)
- Innate immunity and phagocytosis (Yarovinsky)
- Immune responses to parasites (Yarovinsky)
- Computational analysis (Mosmann, Topham)
Minsoo Kim Lab
 Our current knowledge about host immune responses against various virus and bacteria infections stems largely from investigations employing histology or in vitro tissue assays. Until recently, technical difficulties associated with imaging of the immune events within solid organs have limited detection of immune reactions to membranous tissues or superficial regions of non-translucent organs. Dr. Kim is responsible for recent advances in microscopy that made it possible to image immune cell migration and cell-cell interactions in vitro in live cells and also in vivo deep within intact tissues.
Our current knowledge about host immune responses against various virus and bacteria infections stems largely from investigations employing histology or in vitro tissue assays. Until recently, technical difficulties associated with imaging of the immune events within solid organs have limited detection of immune reactions to membranous tissues or superficial regions of non-translucent organs. Dr. Kim is responsible for recent advances in microscopy that made it possible to image immune cell migration and cell-cell interactions in vitro in live cells and also in vivo deep within intact tissues.
- Optogenetically engineered T cells for cancer immunomotherapy
- Inflammatory cues regulating effector CD8 T cell recruitment
- Neutrophil-endothelial interactions and barrier function in sepsis
Alexandra Livingstone Lab
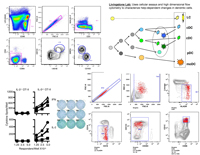 Alexandra Livingstone's lab works on mechanisms of help for CD8+ T cell responses to peptides, tumor & transplantation antigens, and adjuvant-free vaccines, which generally require CD4+ T cell help to generate any detectable primary, memory or protective secondary CD8+ T cell response. We are using computational analysis of polychromatic flow cytometry data (in collaboration with the Mosmann lab), together with multicolor reverse elispot and other assays, to characterize help-dependent, antigen-specific changes in mouse and human dendritic cell subpopulations. We expect to identify novel mechanisms of help that in the absence of overt inflammation are essential for the generation of effective CD8+ T cell responses.
Alexandra Livingstone's lab works on mechanisms of help for CD8+ T cell responses to peptides, tumor & transplantation antigens, and adjuvant-free vaccines, which generally require CD4+ T cell help to generate any detectable primary, memory or protective secondary CD8+ T cell response. We are using computational analysis of polychromatic flow cytometry data (in collaboration with the Mosmann lab), together with multicolor reverse elispot and other assays, to characterize help-dependent, antigen-specific changes in mouse and human dendritic cell subpopulations. We expect to identify novel mechanisms of help that in the absence of overt inflammation are essential for the generation of effective CD8+ T cell responses.
Jim Miller Lab
The identification of CD28 as the primary costimulatory molecule for T cells and of CD80 and CD86 as the ligands for CD28 was a major advance in understanding the regulation of T cell activation. It quickly became clear that CD28 engagement has a dramatic impact on T cell function, through the modulation of a number of different lymphoid cells and a number of different cellular processes. CD28 was shown to enhance initial T cell activation both through upregulation of transcription and the induction of mRNA stability. We found that these two functions of CD28 were mediated by different motifs within the CD28 cytosolic tail domain. Although these functions could be clearly documented they did not account for the full functional activity of CD28. This finding lead us to our current search for other pathways of CD28 regulation and how these pathways impact CD28 function within other aspects of the immune response
Tim Mosmann Lab
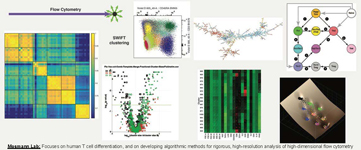 We analyze human T cell differentiation, and the evolution of different T cell subsets during immune responses to vaccination and infection, as well as during pregnancy, atopic dermatitis and cancer. High-dimensional flow cytometry is one of our main tools. To take full advantage of the rich information in large flow datasets, we have developed the SWIFT clustering algorithm in collaboration with Gaurav Sharma in Electrical and Computer Engineering. In conjunction with further analysis tools that we have developed, SWIFT allows high-resolution analysis of datasets, to rigorously analyze known cell types, and to discover unexpected cell populations that change during disease and therapy. These tools are also being applied to test a model of stochastic differentiation.
We analyze human T cell differentiation, and the evolution of different T cell subsets during immune responses to vaccination and infection, as well as during pregnancy, atopic dermatitis and cancer. High-dimensional flow cytometry is one of our main tools. To take full advantage of the rich information in large flow datasets, we have developed the SWIFT clustering algorithm in collaboration with Gaurav Sharma in Electrical and Computer Engineering. In conjunction with further analysis tools that we have developed, SWIFT allows high-resolution analysis of datasets, to rigorously analyze known cell types, and to discover unexpected cell populations that change during disease and therapy. These tools are also being applied to test a model of stochastic differentiation.
Andrea Sant Lab
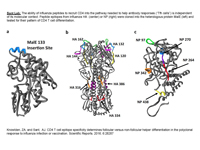 Our laboratory is interested in determining the molecular and cellular events that govern protective immunity to respiratory pathogens, with a particular focus on the cell-mediated response to influenza viruses and vaccines. CD4 T cells convey a multiplicity of diverse effector functions and are critical in orchestrating anti-viral activities by innate and adaptive immune cells. Therefore, our research explores many processes that control CD4 T cell heterogeneity and functionality, including antigen presentation by MHC class II-positive cells in lymphoid and peripheral tissues as well as examining the biochemical composition of licensed vaccines and its importance on the elicitation of protective antibodies to circulating seasonal strains of influenza viruses as well as emerging avian viruses with pandemic potential.
Our laboratory is interested in determining the molecular and cellular events that govern protective immunity to respiratory pathogens, with a particular focus on the cell-mediated response to influenza viruses and vaccines. CD4 T cells convey a multiplicity of diverse effector functions and are critical in orchestrating anti-viral activities by innate and adaptive immune cells. Therefore, our research explores many processes that control CD4 T cell heterogeneity and functionality, including antigen presentation by MHC class II-positive cells in lymphoid and peripheral tissues as well as examining the biochemical composition of licensed vaccines and its importance on the elicitation of protective antibodies to circulating seasonal strains of influenza viruses as well as emerging avian viruses with pandemic potential.
David Topham Lab
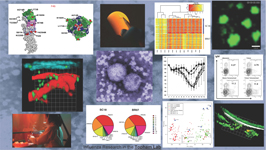 The Topham Lab is focused on Immune Responses to Respiratory Infections and Experimental Vaccines. We have a combined program in basic, clinical, and translational research using animal and computational models of influenza infection and immunity, as well as studies of human immune responses to respiratory infections and experimental vaccines. We have established cutting edge technologies for multidimensional, systems biology based analysis of immune responses. Areas of current research interest include live imaging of T cell responses, motility, and memory in the flu-infected airways, T cell responses to RSV in infants and the elderly, immune system development in infants born prematurely, memory B cell responses to influenza infection and vaccination, genetic and functional diversity in seasonal human influenza, and innate immune regulation by influenza. Our overall goals are to integrate complex data to determine disease mechanisms and predict and mitigate risk from severe disease caused respiratory infections in children and adults.
The Topham Lab is focused on Immune Responses to Respiratory Infections and Experimental Vaccines. We have a combined program in basic, clinical, and translational research using animal and computational models of influenza infection and immunity, as well as studies of human immune responses to respiratory infections and experimental vaccines. We have established cutting edge technologies for multidimensional, systems biology based analysis of immune responses. Areas of current research interest include live imaging of T cell responses, motility, and memory in the flu-infected airways, T cell responses to RSV in infants and the elderly, immune system development in infants born prematurely, memory B cell responses to influenza infection and vaccination, genetic and functional diversity in seasonal human influenza, and innate immune regulation by influenza. Our overall goals are to integrate complex data to determine disease mechanisms and predict and mitigate risk from severe disease caused respiratory infections in children and adults.
Felix Yarovinsky Lab
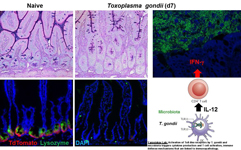 The major interests of our lab involve studying innate immune sensing pathways and their role in regulation of host defense to intracellular pathogens. Our lab is focused on understanding how Toll-like receptors (TLRs) cooperate with other recognition systems to detect protozoan parasites Toxoplasma gondii, Cryprosporidium and malaria and how innate immune recognition regulates IFN-g mediated host defense.
The major interests of our lab involve studying innate immune sensing pathways and their role in regulation of host defense to intracellular pathogens. Our lab is focused on understanding how Toll-like receptors (TLRs) cooperate with other recognition systems to detect protozoan parasites Toxoplasma gondii, Cryprosporidium and malaria and how innate immune recognition regulates IFN-g mediated host defense.
Both Toxoplasma gondii and Cryptosporidium parvum are intracellular protozoan parasites that establish infection through the mucosal tissues after the ingestion of contaminated food products. These pathogens have emerged as an important cause of chronic and fatal disease in immunodeficient individuals, in addition to being investigated as possible triggers of inflammatory bowel disease. Our laboratory aims to understand the mechanisms controlling host-parasite interactions in the gut, a highly specialized environment with distinct structures, epithelial cell types, and innate defense mechanisms required for mucosal host defense.