Single Cell Genomics
Single Cell Genomics
Why Choose Single Cell?

10X Genomics

The 10X Chromium system is a droplet based system capable of capturing up to 20,000 + cells per capture.
Current Capabilities of this platform:
1. Single Cell Gene Expression Profiling
2. Single Cell Immune Profiling
3. Single Cell CNV
4. Single Cell ATAC
5. Single Cell Flex (Cell Fixation, including multiplexing)
6. Single Cell On-Chip Multiplexing
Learn more about the 10X Chromium System
10X Capture Sample Preparation Considerations:
- Capture 1,000 - 10,000 cells with capture rate of ~65% (NextGEM)
Capture 1,000 - 20,000 cells with capture rate of ~70-80% (GEM-X)
- Optimal cell concentrations based on desired targets (GEM-X):
- Target up to 10,000 cells: 700-1200 cells/ul in at least 40ul buffer (minimum 400 cells/ul)
- Target between 10,000 and 20,000 cells: 1300-1600 cells/ul in at at least 40ul buffer (minimum 800 cells/ul)
- Optimal nuclei concentration can range from 300 to 8000 nuclei/ul depending on target number
- Flow sort prior to capture, if possible (isolate particular cell types, remove dead cells)
- Dead cell removal kits can be purchased to improve sample viability. Miltenyi Biotec offers column-based MACS MicroBeads for dead cell remove and other cell depletion or enrichment methods. Please contact us to ensure your enrichment/depletion strategy will be suitable for 10X Genomics single cell chemistry.
- When submitting frozen samples, high quality cells are important. Use freezing media containing 10% DMSO and control the rate of freezing using a freezing container. Greater than 90% viability is recommended. Dead cell removal may be necessary at viabilities below 80%
- Bring GRC single cell suspension with viability >70%. Always keep fresh cells on wet ice to maintain cell viability
- Do NOT use frozen tissue for single-cell suspensions. Instead, nuclei should be isolated from frozen tissue.
Optimal sample quality for Single Cell Submissions:
- Minimal cell debris
- Low cell aggregates
- High cell viability (at least 70%, > 80% is preferred to reach your desired cell target)
- Acceptable Buffers:
- 1X PBS containing 0.04% BSA (Maximum of 1% BSA or up to 10% FBS)
- Buffers should not contain >0.1 mM EDTA or >3 mM magnesium
- Nuclei to be used for gene expression assays must include RNase inhibitor in the resuspension buffer
Nuclei Isolation Services
The GRC offers nuclei isolation to research labs with the following requirements:
- Maximum of 8-10 40um OCT cryosections (or equivalent slices from flash frozen, unembedded tissue).
- Demonstrated protocol can be found here.
- Nuclei quality for submissions:
- Minimal cell debris
- Low nuclei aggregates
- High quality nuclear membranes (healthy nuclei)
- Low intact cell count (% viable cells should be <5%)
If you would like to discuss alternative submission options, please contact us.
GRC resources available to help sample quality
- Sample Filtration: If submitted samples have larger debris the GRC has the ability to filter using cell strainers. Note that cell strainer will NOT be able to remove dead cells or smaller debris
- Sample wash/dilution: For submissions with abundant cells, GRC can wash and/or dilute samples using a 0.04% BSA solution in PBS to help with cell clumping and debris
- Sample concentration: For samples with low concentrations or with smaller debris, the GRC can centrifuge samples using a temperature-controlled swinging-bucket rotor
Cell Hashing, CITEseq, and Feature Barcoding Options:
Options available for multiplexing to lower capture costs and increase cluster identification:
- Cell Hashing: Oligo-tagged antibodies* bind ubiquitously expressed surface proteins allow for unique labeling of samples that can be pooled for a single 10X capture
- CITEseq: Cellular Indexing of Transcriptomes and Epitopes by Sequencing. Antibody* bound oligos act as synthetic transcripts for detection of proteins in quantitative readout.
- Feature Barcoding: CITE/Hashing/other library preparations
*Contact us for more information about our preferred vendors for antibodies.
Newly Released Products from 10X Now Available!
GEM-X:
- Higher Throughput (up to 20,000 cells versus 10,000 for NextGEM)
- Increased Sensitivity (two-fold increase in detected genes, detection of rare transcripts)
- Improved Recovery (Highly viable cells can have a capture rate of up to 80%)
*Please note: NextGEM is being discontinued at the end of 2025 and we will move all projects to GEM-X reagents.
FLEX:
- Profile gene expression for thousands to hundreds of thousands of cells or nuclei with a sensitive probe based method that captures the whole human or mouse transcriptome to detect even low-expressing genes
- Store samples without losing data quality, allowing you to batch samples in the same run and minimize experimental variability
- Fix fresh samples at collection to lock in biological states and preserve fragile cells
- Reduce experimental variability and increase efficiency by batching and multiplexing samples
- Probe-based method for human and mouse. Custom probe options available
- Visit the 10X site about FLEX to learn more.
On-Chip Multiplexing:
- No barcoding prior to capture like with Hashing - reduce time and loss with tagging prior to capture
- Sample multiplexing is accomplished by partitioning up to four samples from each multiplexing set with corresponding gel beads (each containing a unique list of barcodes). A pool of GEMs is generated in a single recovery well for each set. The GEM pool in each recovery well is used to generate cDNA, where DNA derived from a cell shares a common barcode.
- Visit the 10X site about On-Chip Multiplexing to learn more.
Each sample can have up to 5,000 cells for a total of 20,000 cells per capture (or group of 4)
4 samples must be included
For more information about single cell experiment project design or to schedule a 10X capture, please contact us.
Additional testing underway by GRC staff to evaluate other methods such as Illumina Single Cell 3' RNAseq and Parse Biosciences. Contact us to learn more!
Plate-based Methods
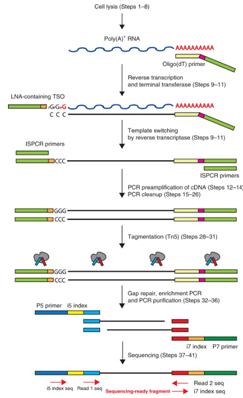
An example of a plate based method is Smart-seq2. In this approach, single cells are placed into each well of a 96-well plate through flow sorting or micro capillary pipetting. With this approach, full length transcripts are captured which allows for detection of splice variants and investigation of SNPs. This method still utilizes the capture of poly(A) RNA and does not indicate strand orientation. Please contact us for more information about this single cell approach.
