Transciptomics of Lung Development
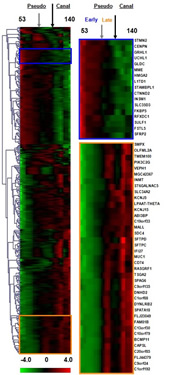
Increased knowledge of the molecular regulation of lung development will help to identify disease mechanisms and therapeutic targets for newborn, pediatric and adult lung diseases. Likewise, attempts to identify lung disease biomarkers will benefit from a greater knowledge of physiological context, including dynamic gene expression levels during normal states such as organ development. Current data strongly supports the concept that developmentally predominant genes and pathways are commonly associated with disease pathogenesis. The lab has significant expertise in the application of genome-wide expression profiling to expand understanding of the regulation of lung formation and its relationship to chronic lung disease pathogenesis. We have demonstrated ability to identify human lung disease expression biomarkers for qualitative and quantitative phenotypes [1]. We have experience in applying data reduction methods to build dynamic models of lung development in the mouse [2-4], which facilitated discovery of gene expression networks associated with biological processes such as the transition to air breathing and alveolar formation. We have generated an expression map of human early fetal lung development, and demonstrated that this complex process can be objectively re-defined according to molecular phases, each composed of anticipated as well as novel pathways [5, 6]. We have applied expression profiling to uncover pathogenic mechanisms in a mouse model of newborn chronic lung disease [7]. Dynamic models of gene expression during development can clearly facilitate the identification of pathways relevant for disease prediction and identification of morbidity-related disease subclasses [8, 9]. Further, we have shown that this approach can facilitate the discovery of lung disease susceptibility genes [10]. We hypothesize that lung physiology (development, homeostasis, disease) is governed by a discrete set of inter-related regulatory networks, such that gaining a better understanding of normal processes will help to identify genes and pathways causal for lung disease. The specific goal of this project is to develop a robust, dynamic model of lung development gene expression, and to resolve this complex process into discrete molecular phases of overlapping biological attributes which will; 1) identify prominent genes and pathways regulating discrete lung developmental processes (e.g., transition to air breathing, alveolarization) relevant to lung disease, and 2) provide a framework to facilitate discovery of pathological changes in gene expression associated with disease phenotypes.