Projects
Projects
- Identification of whole blood mRNA gene expression patterns for distinguishing bacterial from viral and non-infectious respiratory illness (Figure 1)1
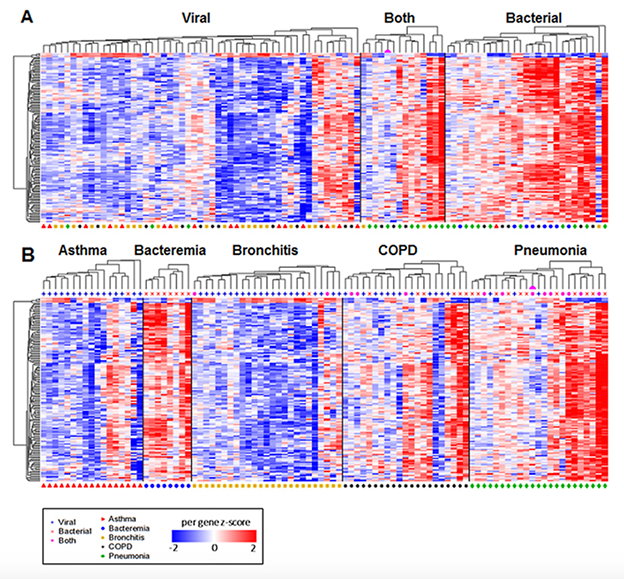
Figure 1. (A) Differential gene expression in patients with viral, bacterial, and co-infections. (B) Gene expression according to clinical syndrome
- Identification of the relationship between nasal epithelial gene expression changes during viral infection and effects on respiratory microbiome as it relates to viral-bacterial co-infection2
- Assessing RSV disease severity relative to viral gene variation, differences in gene expression of peripheral blood CD4, CD8 T cells, and nasal epithelial cells (Figure 2)1,3
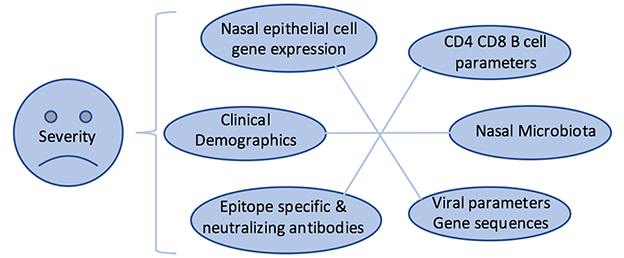
Figure 2. Study design of AsPIRES project illustrating interacting parameters related to RSV disease severity in primary RSV infection.
- Evaluation of the role of nasal microbiota in pathogenesis of RSV in infants1,3
- Clinical trials of anti-RSV antivirals in adults
- Epidemiology studies of clinical impact and population-based incidence of RSV hospitalization in adults
1 In collaboration with Tom Mariani, Department of Pediatrics Transcriptomics Laboratory, and Steve Gill, director of the Genomics Core Laboratory
2 In collaboration with Angela Branche, ID unit
3 In collaboration with Mary Caserta, Department of Pediatrics