Sun Lab
Mechanisms of Defective Viral Genome Generation
We are recruiting now! For anyone who is interested, please contact Dr. Sun!
Defective Viral Genomes (DVGs) are truncated forms of viral genomes that lack the ability to complete replication unless complemented with a homologous helper virus. They are produced during infection with most RNA viruses and they play a critical role in viral pathogenesis. For example, they can reduce the viral load by triggering strong innate immune responses and interfering with full-length viral genome replication. They also facilitate the establishment of viral persistence (Figure 1). While DVGs are important, we don’t know how they arise. The major goal of my lab is to understand the molecular mechanisms and determining factors of DVG generation.
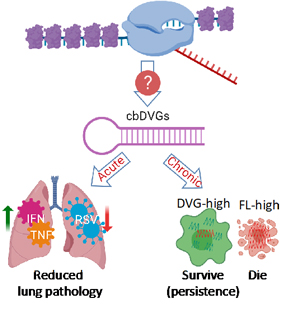
Figure 1: The generation and function of cbDVGs.
We use Respiratory Syncytial Virus (RSV) to study this topic in human, since it is an important human pathogen that causes severe problems in infants, immunocompromised adults, and the elderly and is prone to DVGs. RSV is a single-stranded negative-sense RNA virus, belonging to the pneumoviridae family. It is a leading cause of respiratory illness in children worldwide. Importantly, there are no vaccines or effective treatments available for RSV. RSV generates copy-back type of DVGs (cbDVGs) when the viral polymerase falls off the template and reattaches to the newly synthesized strand, forming a theoretical hairpin loop structure (Figure 1). Previously, we have shown that RSV cbDVGs induce robust antiviral responses, leading to lower viral loads and decreased lung pathology in vivo. Most Excitingly, cbDVGs are observed in RSV infected patients and their presence corelates with higher antiviral responses.
Currently, the lab seeks to understand how cbDVGs are generated during RSV and how manipulation of their generation impacts RSV infection outcome. In order to study DVG generation, we developed an algorithm to survey entire cbDVG populations during infection. To our surprise, we found that despite vast variation in cbDVG species among different RSV infections, cbDVG generation are concentrated in certain genomic hotspots and can be directed by certain mutations within hotspots. We are interested in further understanding the mechanism of how these hotspots regulate cbDVG generation during RSV infection. Furthermore, the lab is also interested in identifying other unknown factors that involve in regulating DVG generation, as we think understanding those factors, and eventually taking advantage of them will allow us to better mitigate virally induced pathology.
Some of the research questions that Sun lab is excited to explore:
- What is the molecular mechanism regulating cbDVG generation in RSV?
- Can we alter viral pathogenesis by manipulating de novo cbDVG generation?
- Are there commonalities in cbDVG generation amongst different viral families, including paramyxoviruses (measles and parainfluenza), pneumoviruses (metapeumoviruses and RSV), and filoviruses (Ebola)?
- What host factors impact cbDVG generation?
- How does the microenvironment influence cbDVG generation?
- What are the mechanism and determining factors for generation of deletion DVGs, another form of DVGs?
Publications
View All Publications- Detection of respiratory syncytial virus defective genomes in nasal secretions is associated with distinct clinical outcomes.; Nature microbiology; Vol 6(5), pp. 672-681. 2021 Apr 01.
- Correction: A specific sequence in the genome of respiratory syncytial virus regulates the generation of copy-back defective viral genomes.; PLoS pathogens; Vol 15(10). 2019 Oct.
- A specific sequence in the genome of respiratory syncytial virus regulates the generation of copy-back defective viral genomes.; PLoS pathogens; Vol 15(4). 2019 Apr.
- Human Genetic Determinants of Viral Diseases.; Annual review of genetics; Vol 51. 2017 Jan 27.
- The innate immune response to RSV: Advances in our understanding of critical viral and host factors.; Vaccine; Vol 35(3). 2017 Jan 11.
- Replication defective viral genomes exploit a cellular pro-survival mechanism to establish paramyxovirus persistence.; Nature communications; Vol 8(1). 2017 Jan 06.
Affiliations
Contact Us
Sun Lab
KMRB 3-9615
601 Elmwood Ave
Rochester, NY 14642