High-Throughput Hyperspectral Two-Photon Fluorescence Imaging
High-Throughput Hyperspectral Two-Photon Fluorescence Imaging
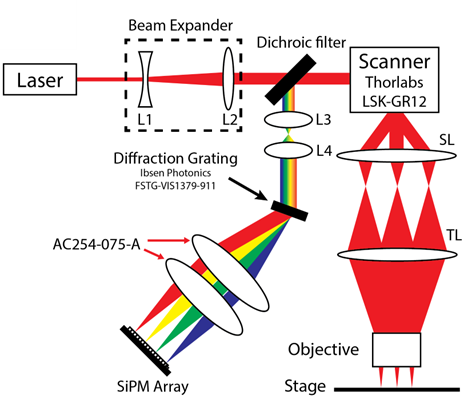
Hyperspectral detection system
The hyperspectral detection system uses a diffraction grating to disperse light by wavelength and then a custom-designed detector array composed of 16 parallel transimpedance amplifiers connected to 16 high dynamic range, single photon sensitive silicon photomultipliers (SiPMs). In contrast to alternative detection technologies such as CMOS arrays, the analog bandwidth of each array element is up to 75 MHz. By leveraging scanner-synchronized stage sampling, the model can image at 50MP/s (800M spectra per second). In contrast to multi-anode PMTs, each SiPM element is completely independent with zero inter-channel crosstalk, and has a saturation power of approximately 40 billion photons per second. As the overall array throughput approaches 1 trillion photons per second, saturation from fluorescence is virtually nonexistent leading to extremely high linearity.
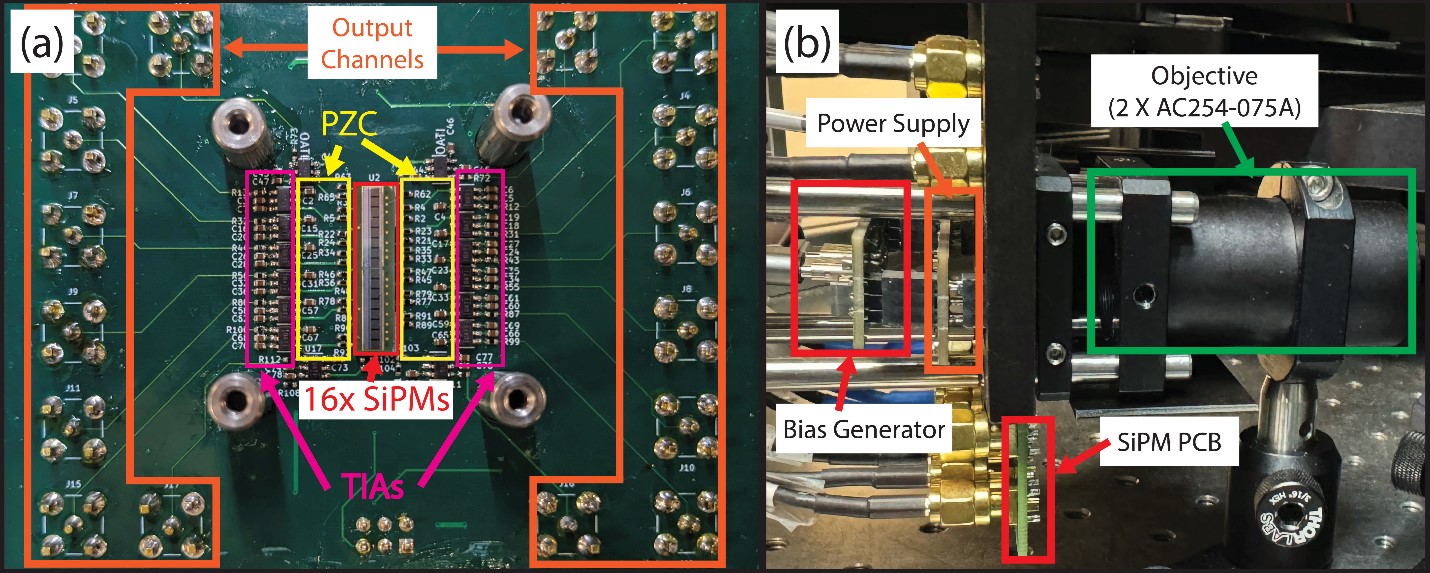
A) Hyperspectral array detector composed of 16 silicon photomultipliers,16 pole-zero cancellation filters, and 16 transimpedance amplifiers. B) Stack up of the high voltage bias generator, analog power supply, SiPM array PCB, and spectrometer objective.
To translate the high-throughput hyperspecial imaging into surgical practice, we developed the hyperPICASSO unmixing algorithm which can identify and separate different spectral features in images using mutual information. Using this algorithm, rapid imaging of tissue components such as keratin is possible, which is otherwise is challenging with conventional stains without substantial delays. We demonstrate here that hyperspectral imaging combined with spectral unmixing can directly discriminate keratinized structures in human squamous cell carcinomas with a simple stain that can be rapidly applied to fresh tissue.
Ongoing work in this project involves improve fluorescence collection efficiency by adopting newer SiPM array with higher fill factor to nondescanned detection utilizing larger array elements, which would enable deep tissue hyperspectral imaging. Miniaturized version of the hyperspecial detector could also adapt to the clinical cart system to perform on-site clinical imaging provides widens the fluorophore choices and enable multiplexing beyond 3-4 channels.
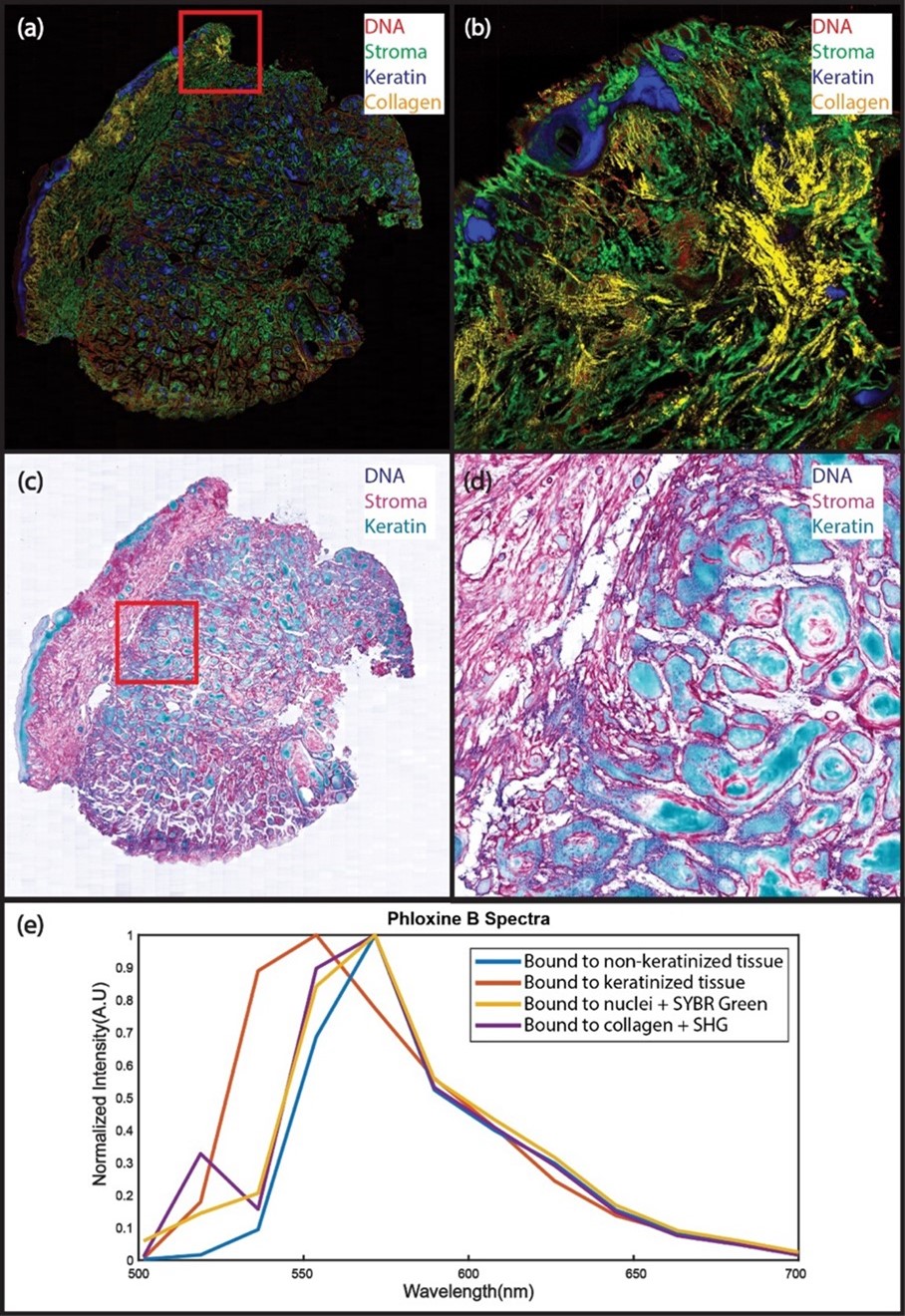
Human surgical margin positive for squamous cell carcinoma labeled with SYBR Green (DNA) and Phloxine B (protein and keratin) imaged at 50 MP/s with hyperPICASSO unmixing. (a): Spectrally unmixed mosaic image with DNA (red), the non-keratin bound PhB (green), keratin-bound PhB (blue), and collagen (Yellow). (b): The enlarged region in B shows keratin-rich regions, such as the epidermis and hair follicles, as well as hallmarks of squamous cell carcinoma, such as keratin pearls. Strong SHG single is from dense connective tissue forming due to a previous biopsy. (c): The mosaic in (a) is rendered as a virtual trichrome stain using the virtual transillumination algorithm (d): The enlarged region shows PhB bound to different tissue types. (e): Spectra from regions in A - D showing the change of PhB emission upon binding to keratinized tissue(orange vs blue), combining emission with SGr on nuclei with additional emission in green (yellow vs. blue), and combining emission on collagens with additional SHG peak(purple vs. blue). Imaging time for the 220 mm2 slide was less than 2 minutes.